The Australian Centre for Ecogenomics (ACE) is a world-leading research hub in the field of ecogenomics and associated bioinformatics.
We provide a focal point for sequence-based genomic data analysis and build capacity to further understand microbial communities and their role in meeting persistent ecological challenges.
Our mission
To explore and understand the microbial world using ecogenomics.
The Australian Centre for Ecogenomics collaborates on active research projects to strengthen our understanding of Australia’s vital microbial communities. We also provide a high-quality sample processing service.
Integrated ecological framework
Our understanding of the microbial biosphere was historically limited because we were unable to grow most microbes in the lab. Culture-independent molecular technologies introduced over the past 30 years have helped us overcome this cultivation bottleneck.
These culture-independent molecular technologies include:
- 16S rRNA gene amplicon profiling of communities
- metagenomics and metatranscriptomics: sequence-based methods that rely on low-cost, high-throughput sequencing of DNA and RNA extracted directly from environmental samples
- cell sorting and whole-genome amplification methods that provide a natural complement to ‘whole’ community characterisation methods
- 16S rRNA-based fluorescence in situ hybridisation (FISH) to image specific groups of microbes
- genome-based classification of bacteria and archaea
- comparative genomics to obtain insights into microbial ecology and evolution.
We define the combination of these techniques as ‘ecogenomics’, which can be used to obtain a holistic overview of microbial communities.
Harnessing microorganisms
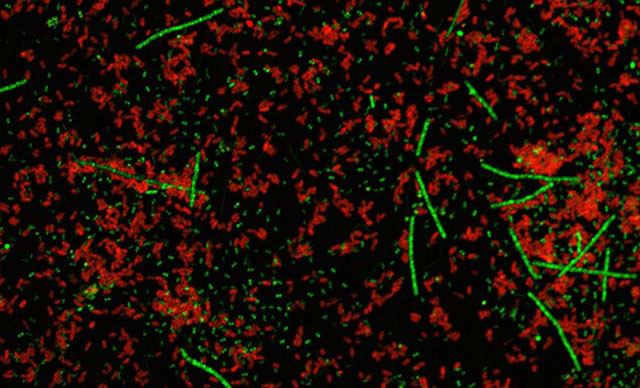
Microorganisms are more than disease agents – they're the most abundant, diverse and ubiquitous forms of life.
In the natural world, microorganisms are globally significant and drive the biogeochemical cycles that sustain life. They directly influence our climate.
Increasingly, researchers harness microorganisms for their diverse metabolic abilities.
Examples include:
- recovering rare earth elements from mine tailings using highly specific microbial proteins
- bioremediation and rehabilitation through microbial activities, for example, acceleratiing soil formation from bauxite residues (red mud)
- using microorganisms to indicate ecosystem health, for example in the Great Barrier Reef
- heat-tolerant enzymes obtained from thermophilic microbes for industrial applications.
With a portfolio that is 80 per cent applied research and 20 per cent fundamental research, we focus on pressing national and international challenges.
Our research themes include:
AI in ecogenomics
Developing insights into protein structure is fundamental as it directly dictates biological functions and interactions. Genetic variations have the potential to disrupt protein function. In humans, these variations are of clinical importance, necessitating the identification of their associations with specific disease phenotypes. In pathogens, variations can lead to resistance, underscoring the need to detect these changes to inform the design of effective drugs that prevent the spread of resistance.
Earth science
The microbiology of earth science ecosystems is coming into greater prominence as Australian industries look for solutions to environmental degradation caused by traditional mining (such as acid mine drainage run-off) and improved methods to extract lower concentrations of metals and rare earth elements.
Host-associated
The community of microorganisms (Bacteria, Archaea, viruses and single-celled Eukaryotes) that function and interact together in a specific location, such as the gastrointestinal tract of an animal, is called the microbiome. In the case of host-associated microbiomes, these interactions extend to the host and are often critical for normal function and development. A dysbiosis, or imbalance of the microbiome, can lead to an increased abundance of microbes that cause disease.
Marine and limnic
Marine and limnic ecosystems underpin life on Earth and are facing increasing pressure from human activity and climate change. Our research encompasses both fundamental and applied understanding of aquatic ecosystems.
Microbial resource management
Microbes are natural recyclers and transformers and can adapt to a variety of food sources due to their large arsenal of encoded functions. We use these microbial abilities to study enzyme-based recycling and bioremediation applications.
Microbial resource management projects
Pathogenomics
We aim to use next-generation DNA sequencing to better understand how medically important Bacteria cause disease, become resistant to antibiotics and disseminate globally.
Tree of life
Metagenomics and single-cell genomics have opened a path to characterising uncultured microbial diversity (so-called 'microbial dark matter'), estimated to represent more than 85 per cent of life on Earth. This new knowledge could provide a greatly improved understanding of how life evolved.
Sequencing services
We analyse sequence-based microbial communities with ecogenomics, an integrated, ecological framework using computational advances and new modelling approaches.
With a range of sequencing options to suit most experimental designs, we:
- deliver comprehensive, high-quality, scientifically valid sample processing
- offer competitive pricing
- apply validated standard operating procedures for processing and analysing samples
- can help with challenging samples and method development
- set no minimum sample number requirement.
Consultancy services
Our microbial resources sampling team offers sample collection, in house DNA sequencing, in house data analysis and report writing, specifically:
- collection of biological materials either by filtration or bulk sample collection
- barcode sequencing of the microbial community to identify community structure
- metagenomic DNA sequencing to identify microbial community structure and function
- informatic comparisons of microbial communities to identify structural and metabolic differences
- expert report writing of microbial community characteristics linked to ecological significance.
Learn more: Analysis of Microbial Communities from Deep Aquifers using DNA Sequence Data (PDF, 352.2 KB)
Contact Dr Paul Evans for more information about our consultancy services: p.evans3@uq.edu.au
Genome Taxonomy Database
The Genome Taxonomy Database is a taxonomic framework that provides a standardised microbial classification based on genome phylogeny.
Our director, Professor Phil Hugenholtz, created the database as a resource for the international research community. It has become a mainstream taxonomic resource with more than 60,000 active users worldwide.
Director
Professor Phil Hugenholtz
Phil Hugenholtz's research profile
Deputy Director
Professor David Ascher
David Ascher's research profile
Group leaders
Associate Professor Scott Beatson
Scott Beatson's research profile
Dr Seweryn Bialasiewicz
Seweryn Bialasiewicz's research profile
Associate Professor Cheong Xin Chan
Cheong Xin Chan's research profile
Dr Paul Evans
Paul Evans's research profile
Researchers
Dr Michaela Blyton
Michaela Blyton's research profile
Dr Kate Bowerman
Kate Bowerman's research profile
Dr Alex de Sa
Dr Akshita Kumar Dhawan
Akshita Kumar Dhawan's research profile
Dr Ashar Malik
Dr Thanh-Binh Nguyen
Thanh-Binh Nguyen's research profile
Dr Paul O'Brien
Paul O'Brien's research profile
Dr Stephanie Portelli
Stephanie Portelli's research profile
Dr Laura Rix
Laura Rix's research profile
Dr Steven Robbins
Steven Robbins's research profile
We also foster the research training of more than 2 dozen HDR students at any given time and are supported by a group of talented professional and technical staff.
Contact us
Get in touch to learn more.
Australian Centre for Ecogenomics
Molecular Biosciences Building, Level 5
The University of Queensland
Brisbane QLD 4072 Australia
Professor Phil Hugenholtz
Director
(07) 3365 3822 p.hugenholtz@uq.edu.au
Mrs Caroline Moniz
Centre Coordinator
(07) 3365 4040 c.moniz@uq.edu.au
Sequencing Team
For more information, including quotes:
