Professor Alan Mark

Primary research interest
Physical and computational chemistry - simulation of biomolecular systems
Group website
About me
After my PhD I held postdoctoral positions at the RSC (ANU) (1987-1988) and at the University of Groningen (1989-1990). In 1990 I moved with W.F. van Gunsteren to ETH Zurich becoming Oberassistant in 1996. In 1998 I was appointed Professor of Biophysical Chemistry, University of Groningen. In 1998 I was awarded the Swiss Ruzicka Prize for research in Chemistry. In 2004 I was awarded an Australian Research Council (ARC) Federation Fellowship and joined The University of Queensland in 2005. In 2011 I was awarded a University of Queensland Vice Chancellor's Senior Research Fellow. I am also an affiliate of the Institute of Molecular Biosciences at UQ and the Australian Infectious Diseases Research Centre.
Research focus and collaborations
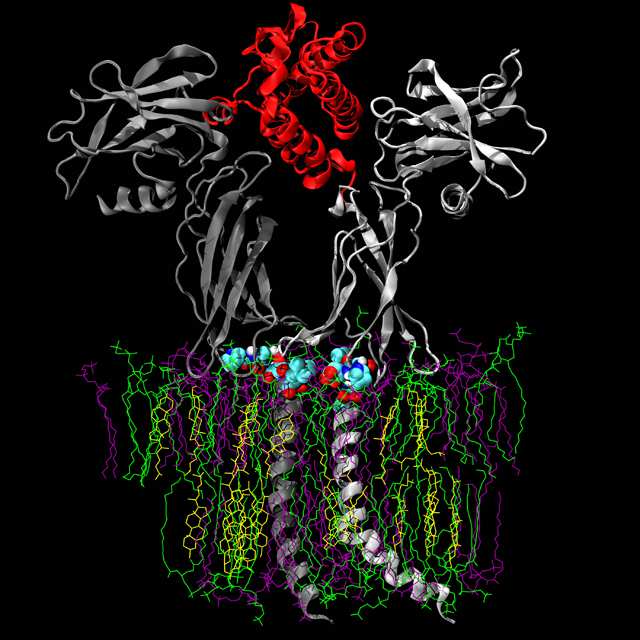
The group focuses on understanding and predicting the macroscopic (experimentally observable) properties of biomolecular systems such as proteins, nucleic acids and lipid aggregates, in terms of the interactions between atoms. In particular, our work concentrates on the development of tools (i.e. simulation software, atomic force fields, theoretical models and experimental techniques) that can be used to understand and predict the physico-chemical basis of interactions and dynamic processes within biomolecular systems. Specific areas of interest include structure prediction, protein and peptide folding, the self-assembly of protein/lipid complexes and the calculation of thermodynamic properties such as ligand binding affinities.
Protein folding
Understanding how proteins fold is one of the grand challenges of modern biology. The failure of proteins to fold correctly is also linked to a range of debilitative diseases including Alzheimer’s disease, BSE and some forms of Type II diabetes where misfolded proteins form destructive aggregates called amyloid fibrils. Research on folding is conducted at multiple levels. Small model systems such as antimicrobial peptides are used to refine force fields and simulation techniques. On a larger scale, we are simulating how multiple copies of certain peptides aggregate in order to understand how amyloid fibrils form.
Cell surface receptors
How the binding of a molecule to an extracellular receptor transfers a signal across the cell membrane or how changes in the environment can activate certain cell surface receptors are both critical question in cell biology. To address such issues we are investigating the mechanism by which low pH triggers the activation of the Dengue E protein, which plays a critical role in the entry of the virus into cells. We are also investigating the structural changes associated with the binding of human growth hormone to the growth hormone receptor.
Membrane protein assembly
Cell membranes are the archetypal self-organised supramolecular structure. Membrane protein complexes also represent a new frontier in structural biology. Using simulations, we are able to directly investigate how bilayers and vesicles form. We are also investigating the assembly of functional structures such as the assembly of anti-microbial peptides into transmembrane pores. This, in turn, is being used to understand the mechanism by which larger complexes form in heterogeneous environments.
Computational drug design
Work in computational drug design is focused in two areas. The development of an automated topology build to provide atomic descriptions of drug-like molecules, the development of novel methods for estimating the free energy of binding and for the refinement of non-standard ligand molecules in X-ray crystal complexes.
International collaborations
The group has a wide range of ongoing collaborations both within Australia and Internationally in Europe and the US, including:
- Prof Wilfred van Gunsteren, ETH, Zurch, Switzerland
- A/Prof Daan Geerke, VU, Amsterdam, The Netherlands
- Prof Gunar Klau, CWI, Amsterdam, The Netherlands
- Prof Siewert-Jan Marrink, University of Groningen, The Netherlands
- Dr Jane Allison, Massey University, New Zealand
Research projects
- Simulating peptide folding and assembly
- Pore-forming peptides as models for protein assembly
- The nucleation and growth of amyloid fibrils
- Mechanism of activation of the human growth hormone receptor
- New methods in drug design
Group members
Dr David Poger, Research Fellow
Funded projects
- NHMRC Project 2015-2018, A New Paradigm for Class I Cytokine Receptor Activation. Andrew Brooks, Michael J. Waters, Alan Mark, Total value of grant: $919,835
- ARC Discovery 2015-2017, Force Fields for Structure Refinement and Computational Drug Design. Alan Mark, Daan Geerke, Gunnar Klau, Total value of grant: $354,000
- ARC Discovery 2013-2015, Membrane proteins: Understanding biological switches, motors and triggers. A E Mark, M L O'Mara, Total value of grant: $946,000
- NHMRC Project 2013-2015, Selective targeting of microbes by peptides of the innate immune system., A E Mark, F Separovic, M-I Aguilar, D Poger, Total value of grant: $606,000
- NHMRC Project 2013-2015, Understanding multidrug resistance in cancer: identification of the substrate and inhibitor binding sites in P-glycoprotein. M L O'Mara, A E Mark, R Callaghan, Total value of grant: $275,000
- NHMRC Project 2012-2014, Structural biology of bacterial lipid IIglycopeptide antibiotic interactions. M A Cooper, A E Mark, Total value of grant: $585,000
- ARC Discovery 2011-2013, Understanding sub-cellular systems at the atomic level. A E Mark, Total value of grant: $300,000
- NHMRC Project 2011-2013, Development of potent and selective blockers of acid sensing ion channels for the treatment of pain. G F King, A E Mark, M A Cooper, P Alewood, L Rash, Total value of grant: $558,000
- ARC Discovery 2008-2010, From structures to systems: A hierarchical approach to understanding sub-cellular components. A E Mark, W F van Gunsteren, S-J Marrink, Total value of grant: $398,000
- ARC Linkage 2008-2010, Development of cryopreservaton for high value provenance collections of recalcitrant plant species used in post-mining restoration., R L Mancera, G J Bryant, A E Mark, S R Turner, P Che, E Bunn, Total value of grant: $807,000
Teaching interests
- Biophysics Major Convenor
- CHEM1022 Chemistry for the Health Professions
- CHEM3011 Advanced Physical Chemistry
Achievements and awards
- 1998 Ruzicka Prize for research in Chemistry
- 2005-2010 Australian Grants Council Federation Fellow.2005-present Extraordinary Professor “Computer simulation of biological molecules,” University of Groningen, The Netherlands.
- 2012-present University of Queensland Vice Chancellor's Senior Research Fellow.
- 2012 The Association of Molecular Modellers of Australasia (AMMA) medal.
- 2013-2015 Chair of the Merit Allocation Committee for the National Cyber Infrastructure
- 2013-2015 Australian Grants Council Discovery Outstanding Researcher Award (DORA)
Featured publications
- Brooks, Andrew J., Dai, Wei, O’Mara, Megan L., Abankwa, Daniel, Chhabra, Yash, Pelekanos, Rebecca A., Gardon, Olivier, Tunny, Kathryn A., Blucher, Kristopher M., Morton, Craig J., Parker, Michael W., Sierecki, Emma, Gambin, Yann, Gomez, Guillermo A., Alexandrov, Kirill, Wilson, Ian A., Doxastakis, Manolis, Mark, Alan E. and Waters, Michael J. (2014) Mechanism of activation of protein kinase JAK2 by the growth hormone receptor. Science, 344 6185: 1249783.1-1249783.12. doi:10.1126/science.1249783
- Poger, David and Mark, Alan E. (2014) Activation of the epidermal growth factor receptor: a series of twists and turns. Biochemistry, 53 16: 2710-2721. doi:10.1021/bi401632z
Researcher biography
We use computer based modelling techniques to understand and predict the the structural and dynamic properties of (bio)molecules including proteins and lipid aggregates.
Born in 1961, I obtained a BSc (Hon 1) at the University of Sydney in 1982. I obtained my PhD in 1986 from the John Curtin School of Medical Research, Australian National University (ANU), on the "Binding Responses Associated with Self-Interacting Ligands: Studies on the Self-Association and Receptor binding of Insulin". After holding postdoctoral positions at the ANU, University of Groningen, The Netherlands and the Federal Institute of Technology (ETH), Zurich, Switzerland I was appointed Professor of Biophysical Chemistry (Molecular Simulation) University of Groningen, in 1998. In 1998 I also received the Swiss Ruzicka Prize for research in Chemistry for work on simulating peptide folding. In 2004 I was awarded an ARC Federation Fellowship and in February 2005 an honorary chair (Bijzonder Hoogleraar) at the University of Groningen, The Netherlands. I have given over 90 invited lectures at conferences and academic Institutions around the world as well as at a range of summer and winter schools on advanced simulation techniques.
In my research I have performed pioneering simulations of a variety of important biological phenomena, including some of the first atomic simulations of protein unfolding and the first simulations of reversible peptide folding in a realistic environment. In recent years my group performed some of the first atomic and near atomic simulations of the spontaneous aggregation of surfactant and lipid systems into micelles, bilayers and vesicles. These have enabled us, amongst other things, to elucidate the mechanism by which pores are induced within biological membranes in unprecedented detail. Over the last decade I have been intimately involved in the development of the GROMOS force field which is specifically tuned for protein and peptide folding simulations and as well as the development of models for a range of solvents including methanol and trifluoroethanol. I have also been responsible for the development of methodology for the calculations of the thermodynamic properties of biomolecular systems such as free energies of binding and hydration, as well as estimating entropic effects from simulations. Most recently, I have been responsible for the development of novel approaches to promote structure formation in protein folding simulations that can be used for the refinement of protein structures generated by ab initio or by homology methods. Finally, I am associated with two, internationally recognised, (bio)molecular simulation packages the GROningen Molecular Simulation library (GROMOS) and the GROningen Machine for Chemical Simulations (GROMACS).
